0
Samples analysed
0
Target species assays
0
Sites surveyed

What is evironmental DNA (eDNA)?
All species shed traces of DNA into the environment through skin, faeces, blood, mucus, sperm, and other biological material. This is known as environmental DNA (eDNA). By analysing small fragments of this DNA in water or soil samples, we can detect species across large or hard-to-reach areas without the need to physically see or capture them.
Results are delivered in a report with expert interpretation to help you understand what was detected and what it means.
We ensure high-quality data:
-
Our eDNA metabarcoding data is reviewed by ecologist experts on each particular taxon (fish, amphibians, invertebrates, etc) to minimise species misassignments
-
Our targeted eDNA assays are developed following international guidelines, and our data is always Sanger-sequenced to minimise false positives
We advise whether further field surveys are recommended. Where relevant, we highlight biosecurity concerns or whether the detection supports recovery or conservation planning.
High-quality data

Results: expert interpretation for management, biosecurity and conservation
Samples collected in the field are sent to our lab, where DNA is extracted and analysed. All lab protocols are designed to maximise eDNA detection and ensure accurate, reliable results.
Our team uses specialised equipment to extract eDNA directly from large volumes of water, so field teams don’t need to filter samples. We’ve also developed preservation methods that don’t require refrigeration, making it easier to collect and store samples in remote areas for later analysis.
Analysis in the lab
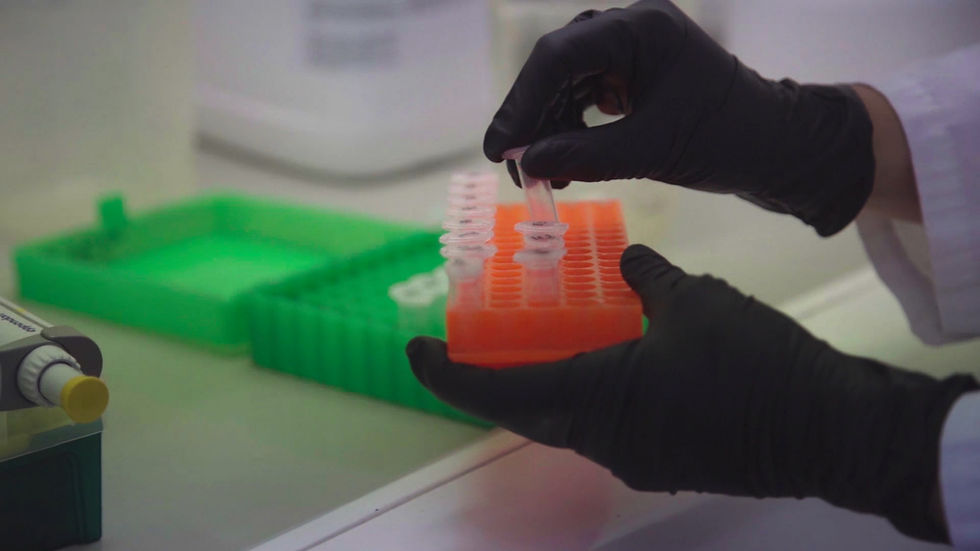
Our lab: advanced analysis and preservation methods
Our eDNA kits
Each eDNA kit is designed by our scientists to maximise detection, by either targeting a specific species (like yellow crazy ants) or a broader group (such as all fish or bacteria) using metabarcoding.
We also adapt sampling methods to suit different environments, from fast-flowing rivers to estuaries and the open ocean.
We provide training so that anyone, including community groups and Traditional Owners, can use these kits to collect water or soil samples. Their simplicity supports widespread, community-led monitoring across large and remote areas.

eDNA kits: how they work and who collects samples
We are northern Australia’s leading eDNA experts
Using advanced techniques in our lab, our scientists have developed targeted eDNA methods to detect and monitor invasive and endangered species. We’ve contributed to biosecurity surveillance, environmental impact assessments, tracking fish communities in Australian waterways, locating threatened species, and advancing eDNA science itself, while working with Traditional Owners, community groups, governments, councils and regional bodies.
Locating and protecting threatened species
We have advanced eDNA technology to detect threatened species, such as frogs, turtles, and freshwater fish, without needing to see or capture them. This is highly effective at surveying large or remote areas, identify hard-to-find species, and minimise disturbance to sensitive habitats. It helps conservationists, land managers, and Traditional Owner groups make informed decisions and respond more effectively to species decline.


Finding ‘missing’ frogs using eDNA
TropWATER scientists developed a method to detect endangered frog populations by tracing their DNA in waterways – even when the frogs are more than 20 kilometres upstream. This approach is helping locate elusive species once thought to be lost, and is transforming how threatened amphibians are surveyed across northern Australia.
We rediscovered the Irwin turtle
TropWATER scientists used environmental DNA (eDNA) to confirm the presence of the Irwin’s turtle in the lower Burdekin River – the first formal record in more than 25 years. This rediscovery, made possible through water sampling across three river catchments, challenges earlier assumptions about the species’ survival in turbid waters and shows the power of eDNA to detect elusive species in remote and crocodile-prone areas.
Strengthening community-led efforts
We work closely with community groups, Traditional Owners, Indigenous Rangers, and local councils to make eDNA science more accessible and locally relevant. By supporting community-led monitoring and offering training in sampling techniques, we’re helping build capacity on the ground. These partnerships strengthen local knowledge, improve long-term data collection, and create shared ownership in the protection of northern Australia’s waterways and biodiversity.


Tracking cane toads
We developed a highly sensitive eDNA method that can detect the presence of a single cane toad from just a brief visit to a small waterbody, even when toads only stayed for five minutes. In partnership with the Torres Strait Regional Authority, this technique is now helping Indigenous Rangers monitor cane toads across the Torres Strait, providing a practical tool to detect early invasions and support local biosecurity efforts.
Monitoring fish communities
We’ve worked with local groups like OzFish, Creekwatch, and Traditional Owners to monitor fish communities in creeks and rivers using eDNA. By training community members to collect water samples, species can be detected without relying on traps or nets. This approach has helped communities in Townsville, the Burdekin, and the Herbert region better understand the fish living in their waterways and contribute to long-term monitoring and management.
Biosecurity for invasive species
Our eDNA technology allows early detection of invasive species, like yellow crazy ants and Varroa mites. It enables rapid biosecurity response and management to stop their spread and be used as a surveillance tool. We can deliver a fast, cost-effective methods to improve biosecurity by detecting low-density and hidden species over large areas.


Invasive ants on Reef Islands
Invasive ants like electric ants, fire ants, and yellow crazy ants pose a serious biosecurity risk to Great Barrier Reef islands, threatening native wildlife such as bird chicks and turtle hatchlings. TropWATER researchers, working with Traditional Owner groups, used eDNA to detect these species by analysing soil samples for their unique DNA. The project developed and field-tested targeted detection methods to support early intervention and protect island ecosystems.
Screening for Varroa mites
Varroa mites are one of the most serious biosecurity threats to Australia’s honeybee industry, weakening bee populations and putting pollination-dependent crops and ecosystems at risk. To support early intervention, TropWATER researchers have developed reliable eDNA protocols to detect Varroa mites in honeybee populations. This work has also contributed to portable diagnostic tools for rapid, on-site screening — improving response times and strengthening national efforts to contain outbreaks.
Advancing eDNA science
We have advanced eDNA methods for science to detect both individual species and broader groups of species using genetic markers, with both processes requiring detailed genetic knowledge and rigorous lab testing. To accurately detect rare or invasive species, our team designs species-specific primers. We have also advanced metabarcoding methods, which are a newer eDNA technique that detects many species at once by analysing shared genetic markers in samples. This provides a broader view of biodiversity and how ecological communities change over time.


First to detect invertebrates
TropWATER was among the first to successfully detect terrestrial invertebrates using eDNA, demonstrating that species like yellow crazy ants could be identified from water samples — even when infestations were located hundreds of metres from the source. This early breakthrough expanded the scope of eDNA beyond aquatic species and paved the way for new applications in terrestrial biosecurity and pest surveillance.
Tracking cane toads
Metabarcoding improves soil health
TropWATER scientists are expanding the use of eDNA metabarcoding to study how microbes and invertebrates cycle nutrients in agricultural soils. This technique detects multiple microbial and invertebrate organisms at once, helping reveal how different farming practices influence nutrient retention, leaching, and runoff. The research is advancing eDNA science while supporting landholders to improve soil health, reduce fertiliser losses, and improve water quality.
Explore our work
Use our map below to explore different projects and target species.
eDNA FAQ
Looking for more information? Here are answers to some commonly asked questions about eDNA sampling and analysis.
Still have questions? Contact us about our eDNA lab and kits.
Target species detection uses a highly specific test, called an assay, to look for the DNA of one species, such as the endangered armoured frog or the invasive yellow crazy ant. Every field sample collected is scanned for this specific assay.
Metabarcoding scans for a broad group of species (like all fish or all bacteria) by targeting a gene section they all share. It gives you a snapshot of the biodiversity present in your sample. Both approaches rely on reference databases of known DNA sequences.
Metabarcoding is ideal when you want to understand what communities of species live in an environment. It’s used to assess biodiversity in rivers, oceans, or soil, track changes in ecosystems over time, or study microbes involved in soil and water health. It's also useful when you're not sure what species you're looking for. The most common species tend to appear most in the results because they collectively shed more eDNA, but rare species can still be detected.
No – while metabarcoding identifies a broad group of species with a shared gene in a sample, identifying all species from all groups is very challenging for technical and practical reasons.
Technical reasons include the sampling design (eDNA from some species may not be captured due to low abundance, low shedding rates, or suboptimal sampling) and PCR efficiency (DNA from some species will be more easily amplified than others). Metabarcoding assays are designed to focus on a targeted group of species (such as fish, corals, or microbes) so species outside that group will not be detected. Practical limitations involve reference databases; DNA sequences must be matched to known references, and not all species have reference DNA available.
Targeted detection is used when you are searching for one specific species. Since it is the most sensitive eDNA method of detection, it is useful when trying to locate an endangered or invasive species because the presence or absence of an individual of that species could have implications for conservation or biosecurity. Targeted detection is also valuable for locating species whose characteristics, like camouflage or remaining hidden, make them difficult to observe directly.
The way eDNA samples are collected and analysed has revolutionised how we can detect species across a range of environments. Sampling is quick and easy, allowing for screening of large areas, and it does not require expertise. Detection is also indirect, as it doesn’t require sighting the species directly, so it can be used to find rare/cryptic species or provide a broad snapshot of whole communities in an ecosystem. This can be a powerful tool for early detection of invasive species, before sightings occur.
No, not all eDNA labs operate at the same level. Our lab:
-
Delivers both commercial and research services, while developing new field and lab methods to advance eDNA science and ensure robust data.
-
Targeted eDNA: We design assays to international best practice guidelines, test them against our large fish and reptile tissue collection, and validate all positive detections through Sanger sequencing. This guarantees that all positive detections have been scrutinised.
-
Metabarcoding: Our data is reviewed by expert ecologists with knowledge of Australian biodiversity in each taxonomic group (e.g., fish, vertebrates, invertebrates) to reduce false positives from taxonomic misassignments.
-
Training: We provide training in eDNA methods, covering theory, sampling techniques, and data interpretation.
-
Yes. Soil and water samples can be collected from terrestrial, freshwater, coastal, and marine environments. We can design a sampling plan tailored to your project and research questions.
Species from across a wide range of taxonomic groups can be detected using eDNA analysis, including bacteria, viruses, fungi, and plants as well as mammals, birds, fish, invertebrates, and more. Although some organisms naturally shed more DNA than others, even rare species can be detected using appropriate eDNA protocols.
No – our sampling kits have been specifically designed to be used in tropical remote areas of northern Australia. Sampling simply consists of filling up a jar with water and pouring it into another jar containing the preservative buffer. This means that there is no need to specialised equipment, not even a fridge or ice, since the samples do not have to be kept cool.
Water samples can last for between six weeks to three months, depending on temperatures. We have trialled the efficiency of the preservative buffer and it can keep eDNA intact in temperatures as high as 60°C for at least six weeks and ambient temperatures for at least three months.
-
Project goals: The TropWATER eDNA team meets with you to discuss project objectives.
-
Sample collection: Water, soil, or air samples are taken using a field kit provided by TropWATER.
-
DNA extraction: The sample is processed in a lab to extract any DNA present.
-
Analysis for metabarcoding or target species: The target gene sequences for a taxon/organism are identified, matched to databases, and interpreted based on the species you’re targeting or group you’re assessing.
-
Results: A report is provided that includes a detailed account of the methods used, results on species detection, and recommendations.
-















